UAS vectors:
| Name | Features | Components | Comment | Depository ID | Sequence | Reference |
| pUA | 10X UAS version of pUAST-attB | 10X UAS; hsp70 promoter/5’UTR; SV40 early polyA | attB only | Addgene 58372 | download | Han et al., 2014 |
| pACU | original pUAST with attB | 5X UAS; hsp70 promoter/5’UTR; SV40 early polyA | recommended for low expression | Addgene 58373 | download | Han et al., 2011 |
| pACUH | 10X UAS version of pACU | 10X UAS; hsp70 promoter/5’UTR; SV40 early polyA | recommended for intermediate expression | Addgene 58374 | download | Sapar et al., 2018 |
| pACU2 | high expression with HIH cassette | 5X UAS; hsp70 promoter/5’UTR; synthetic intron; His2Av polyA | possible leaky expression induced by enhancers near the insertion site | Addgene 31223 | download | Han et al., 2011 |
| pIHEU | insulated version of pACU2 | 5X UAS; hsp70 promoter/5’UTR; synthetic intron; His2Av polyA; gypsy insulators | recommended for high expression without Gal4-independent leaky expression | Addgene 58375 | download | Han et al., 2011 |
- Except for pUA, all plasmids were constructed in the dual-transformation backbone pAC (attB CaSpeR) that contains attB, P-element, and mini-white.
- Expression level: pIHEU ≈ pACU2 > pACUH ≈ pUA > pACU
Gateway Destination vectors
| Name | Purpose | Components | Comment | Depository ID | Sequence | Reference |
| pDEST-HemmarR | For making enhancer-driven CD4-tdTomato marker | hsp70 promoter/5’UTR; zeste intron; CD4-tdTomato CDS; His2Av polyA | original Hemmar vector | Addgene 31222 | download | Han et al., 2011 |
| pDEST-HemmarG | For making enhancer-driven CD4-tdGFP marker | hsp70 promoter/5’UTR; zeste intron; CD4-tdGFP CDS; His2Av polyA | original Hemmar vector | Addgene 31221 | download | Han et al., 2011 |
| pDEST-HemmarR2 | For making enhancer-driven CD4-tdTomato marker | DSCPm; zeste intron; CD4-tdTomato CDS; His2Av polyA | Modified Hemmar vector that matches the expression of FlyLight Gal4 constructs better | Addgene 112813 | download | Poe et al., 2018 |
| pDEST-HemmarG2 | For making enhancer-driven CD4-tdGFP marker | DSCPm; zeste intron; CD4-tdGFP CDS; His2Av polyA | Modified Hemmar vector that matches the expression of FlyLight Gal4 constructs better | Addgene 112814 | download | Poe et al., 2018 |
| pDEST-HemmarI2 | For making enhancer-driven CD4-IFP2.0 marker | DSCPm; zeste intron; CD4-IFP2.0-T2A-HO1 CDS; His2Av polyA | Modified Hemmar vector that matches the expression of FlyLight Gal4 constructs better | Addgene 112815 | download | Poe et al., 2018 |
| pDEST-APIGH | For making enhancer-driven Gal4 | hsp70 promoter/5’UTR; zeste intron; Gal4 CDS; His2Av polyA | Addgene 112804 | download | Han et al., 2014 | |
| pDEST-APIC-Flp1 | For making enhancer-driven Flp1 | hsp70 promoter/5’UTR; zeste intron; Flp1 CDS; His2Av polyA | Flp1 is a high activity Flp with D at AA5 | Addgene 112806 | download | Poe et al., 2017 |
| pDEST-APIC-LexAGAD | For making enhancer-driven LexAGAD | hsp70 promoter/5’UTR; zeste intron; LexAGAD CDS; His2Av polyA | Addgene 112807 | download | Poe et al., 2017 | |
| pDEST-APIC-Cas9 | For making enhancer-driven Cas9 | hsp70 promoter/5’UTR; zeste intron; Cas9 CDS; His2Av polyA | Addgene 121657 | download | Poe et al., 2018 | |
| pDEST-APLO | For making 13X LexAop2-driven transgene | 13X LexAop2; hsp70 promoter/5’UTR; synthetic intron; His2Av polyA | Addgene 112805 | download | Poe et al., 2017 |
- All plasmids were constructed in pAIPC (attB P-element insulated CaSpeR), a dual-transformation backbone that contains attB, P-element, mini-white, and two gypsy insulators flanking the transgene unit. All contain ccdB cassette (attR1/attR2) for introducing DNA fragments in entry vectors through Gateway L/R reactions.
- DSCPm: modified Drosophila synthetic core promoter based on hsp70 promoter/5’UTR, containing Inr, MTE, DPE, in addition to TATA box
CRISPR Reagents
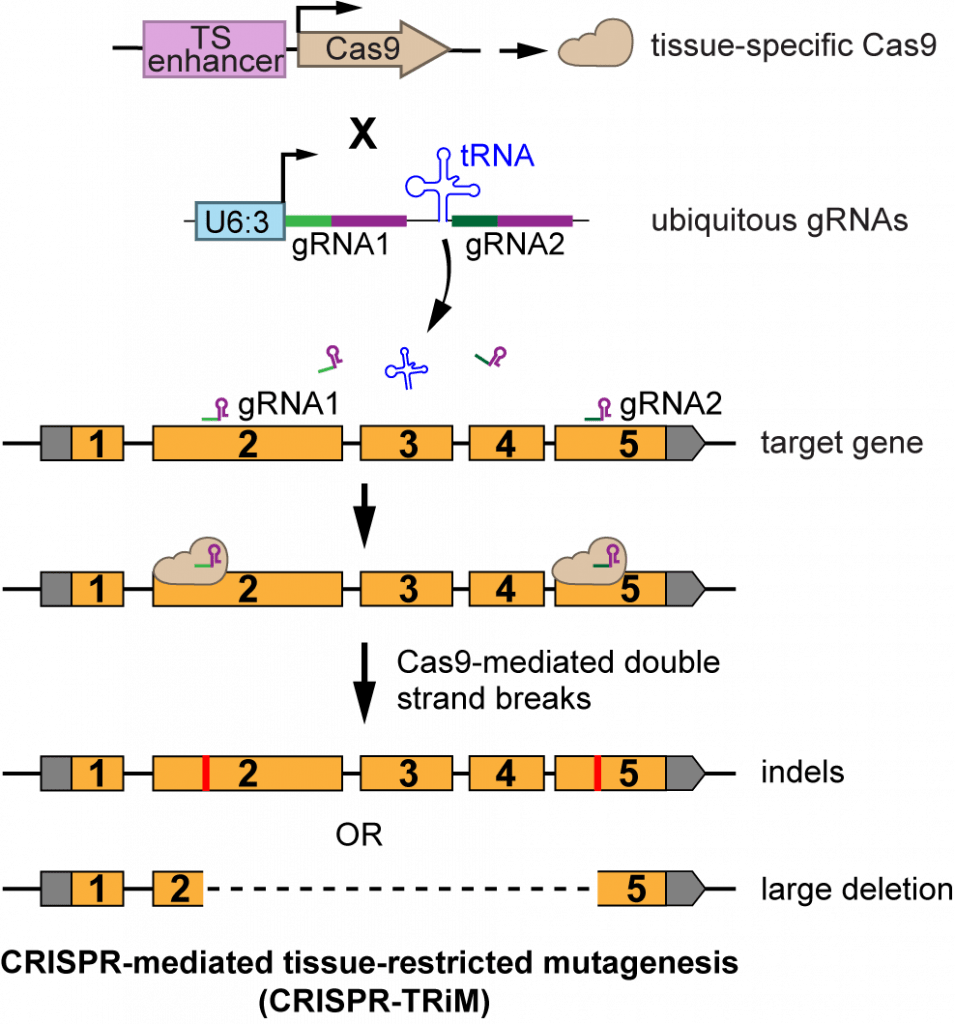 |
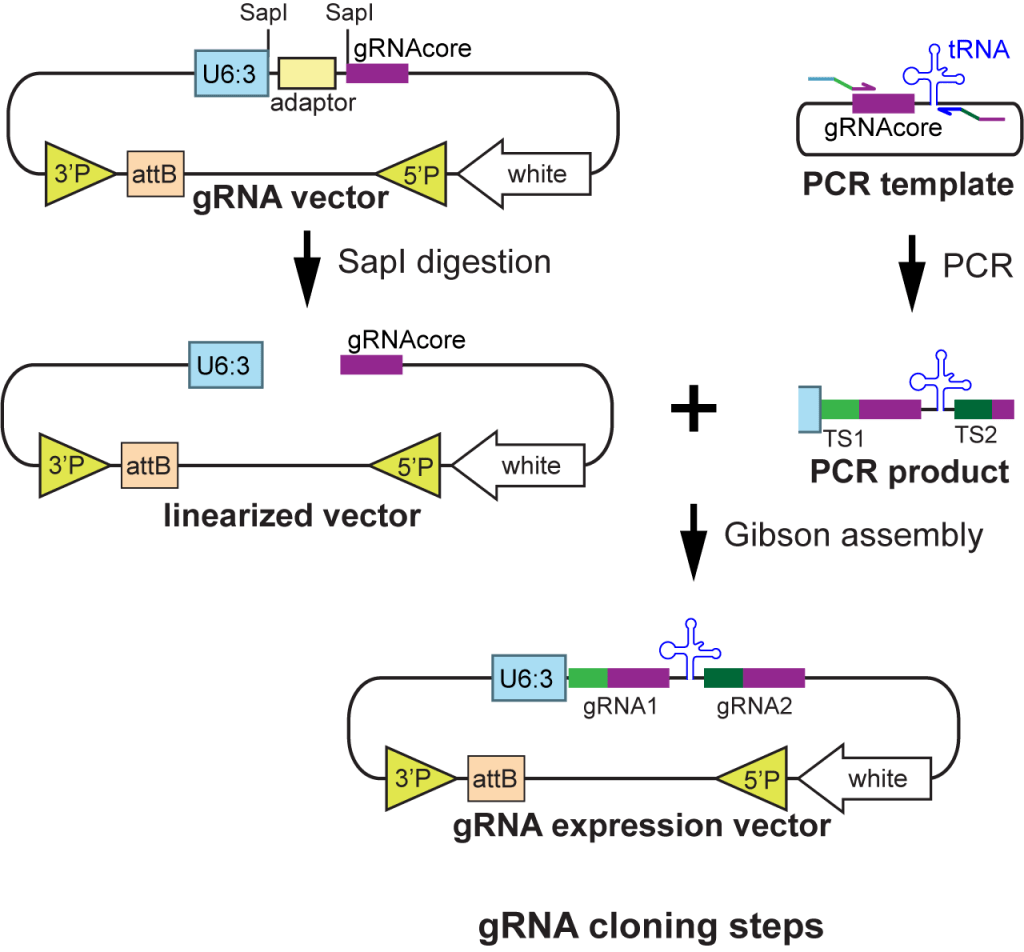 |
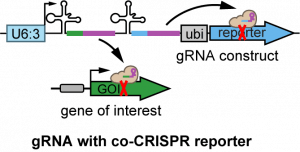 |
Mosaic Analysis by gRNA-induced Crossing-over (MAGIC)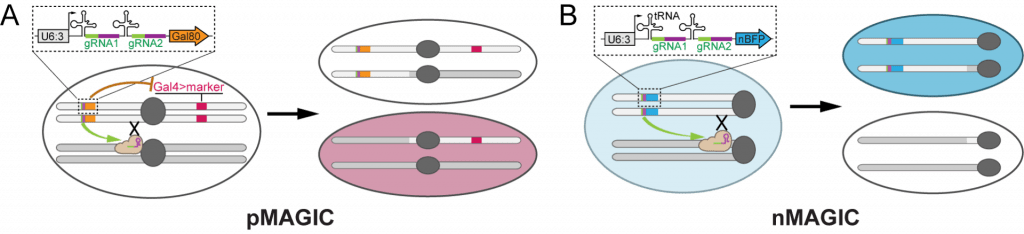
gRNA cloning vectors for CRISPR-TRiM and MAGIC:
| Name | Purpose | Components | Comment | Depository ID | Sequence | Reference |
| Tissue-specific CRISRPR (CRISPR-TRiM) | ||||||
| pAC-U61-SapI | For making dual- and triple-gRNAs with separate U6 promoters; original gRNA scaffold | U6:1 promoter; SapI adaptor for cloning inserts by Gibson Assembly; original gRNA scaffold | Old generation; not recommended | Addgene 112808 | download | Poe et al., 2018 |
| pTGC-U6.2 | PCR template vector to use together with pAC-U61-SapI | gRNAcore(original)-U6:2 | Addgene 112809 | download | Poe et al., 2018 | |
| pTGC-U6.3 | PCR template vector to use together with pAC-U61-SapI | gRNAcore(original)-U6:3 | Addgene 112810 | download | Poe et al., 2018 | |
| pAC-U63-tgRNA-Rev | For making multiplexed gRNAs separated by tRNAGly spacers; gRNA(F+E) scaffold | U6:3 promoter; tRNAGly ; SapI adaptor for cloning inserts by Gibson Assembly; gRNA(F+E) scaffold | Old generation; not recommended | Addgene 112811 | download | Poe et al., 2018 |
| pMGC |
PCR template vector to use together with pAC-U63-tgRNA-Rev, pAC-U63-tgRNA-nlsBFP, or pAC-U63-tgRNA-Gal80 |
gRNAcore(F+E)-tRNAGly | Addgene 112812 | download | Poe et al., 2018 | |
| pTR(EF)-tRNA(Q) |
PCR template vector to use together with pAC-U63-tgRNA-Rev, pAC-U63-tgRNA-nlsBFP, or pAC-U63-tgRNA-Gal80 |
gRNAcore(F+E)-tRNAGln | Addgene 170517 | download | Koreman et al., 2021 | |
| pAC-U63-gRNA2.1 | For making multiplexed gRNAs separated by tRNAGln spacers; gRNA(2.1) scaffold | U6:3 promoter; SapI adaptor for cloning inserts by Gibson Assembly; gRNA(2.1) scaffold | Highest efficiency in somatic tissues; recommended for tissue-specific CRISPR | Addgene 170512 | download | Koreman et al., 2021 |
| pAC-U63-QtgRNA2.1-BR | For making multiplexed gRNAs with BFP co-CRISPR reporter; also serving as the PCR template vector for cloning in pAC-U63-gRNA2.1 | U6:3 promoter; SapI adaptor for cloning inserts by Gibson Assembly; gRNA(2.1) scaffold; tRNAGln; gRNA-BFP; ubi-nlsBFP | Recommended for labeling mutant cells by the loss of nuclear BFP | Addgene 170513 | download | Koreman et al., 2021 |
| pAC-U63-QtgRNA2.1-8R | For making multiplexed gRNAs with Gal80 co-CRISPR reporter; also serving as the PCR template vector for cloning in pAC-U63-gRNA2.1 | U6:3 promoter; SapI adaptor for cloning inserts by Gibson Assembly; gRNA(2.1) scaffold; tRNAGln; gRNA-Gal80; ubi-Gal80 | Recommended for labeling mutant cells by Gal4-driven fluorescent markers | Addgene 170514 | download | Koreman et al., 2021 |
| pAC-CR7T-gRNA2.1-nlsBFP | For making dual-gRNAs with separate promoters; gRNA(2.1) scaffold | CR7T promoter; SapI adaptor for cloning inserts by Gibson Assembly; gRNA(2.1) scaffold; ubi-nlsBFP | Highest efficiency in the germline; recommended for germline mutagenesis and HDR-mediated knock-in | Addgene 170515 | download | Koreman et al., 2021 |
| pGC(2.1)-U6.3 | PCR template vector to use together with pAC-CR7T-gRNA2.1-nlsBFP | gRNAcore(2.1)-pU6.3 | Addgene 170516 | download | Koreman et al., 2021 | |
| Mosaic Analysis by gRNA-induced Crossing-over (MAGIC) | ||||||
| pAC-U63-tgRNA-nlsBFP | For making nMAGIC gRNA-marker transgene | U6:3 promoter; tRNAGly ; SapI adaptor for cloning inserts by Gibson Assembly; gRNA(F+E) scaffold; ubi-nlsBFP | Recommended for labeling mosaic clones by the loss of nuclear BFP | Addgene 169029 | download | Allen et al., 2021 |
| pAC-U63-tgRNA-Gal80 | For making pMAGIC gRNA-marker transgene | U6:3 promoter; tRNAGly ; SapI adaptor for cloning inserts by Gibson Assembly; gRNA(F+E) scaffold; ubi-Gal80 | Recommended for labeling mosaic clones by by Gal4-driven fluorescent markers | Addgene 169030 | download | Allen et al., 2021 |
- All gRNA vectors were constructed in the dual-transformation backbone pAC (attB CaSpeR) that contains attB, P-element, and mini-white.
Reference:
Allen, S.E., Koreman, G.T., Sarkar, A., Wang, B., Wolfner, M.F., and Han, C., 2021. Versatile CRISPR/Cas9-mediated mosaic analysis by gRNA-induced crossing-over for unmodified genomes. PLoS Biol 19, e3001061. https://doi.org/10.1371/journal.pbio.3001061.
Han, C., Jan, L.Y., and Jan, Y.N., 2011. Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc Natl Acad Sci U S A 108, 9673-9678. https://doi.org/10.1073/pnas.1106386108.
Han, C., Wang, D., Soba, P., Zhu, S., Lin, X., Jan, L.Y., and Jan, Y.N., 2012. Integrins regulate repulsion-mediated dendritic patterning of drosophila sensory neurons by restricting dendrites in a 2D space. Neuron 73, 64-78. https://doi.org/10.1016/j.neuron.2011.10.036.
Han, C., Song, Y., Xiao, H., Wang, D., Franc, N.C., Jan, L.Y., and Jan, Y.N., 2014. Epidermal cells are the primary phagocytes in the fragmentation and clearance of degenerating dendrites in Drosophila. Neuron 81, 544-560. https://doi.org/10.1016/j.neuron.2013.11.021.
Koreman, G.T., Xu, Y., Hu, Q., Zhang, Z., Allen, S.E., Wolfner, M.F., Wang, B., and Han, C., 2021. Upgraded CRISPR/Cas9 tools for tissue-specific mutagenesis in Drosophila. Proc Natl Acad Sci U S A 118. https://doi.org/10.1073/pnas.2014255118.
Poe, A.R., Tang, L., Wang, B., Li, Y., Sapar, M.L., and Han, C., 2017. Dendritic space-filling requires a neuronal type-specific extracellular permissive signal in Drosophila. Proc Natl Acad Sci U S A 114, E8062-E8071. https://doi.org/10.1073/pnas.1707467114.
Poe, A.R., Wang, B., Sapar, M.L., Ji, H., Li, K., Onabajo, T., Fazliyeva, R., Gibbs, M., Qiu, Y., Hu, Y., et al., 2019. Robust CRISPR/Cas9-Mediated Tissue-Specific Mutagenesis Reveals Gene Redundancy and Perdurance in Drosophila. Genetics 211, 459-472. https://doi.org/10.1534/genetics.118.301736.
Sapar, M.L., Ji, H., Wang, B., Poe, A.R., Dubey, K., Ren, X., Ni, J.Q., and Han, C., 2018. Phosphatidylserine Externalization Results from and Causes Neurite Degeneration in Drosophila. Cell Rep 24, 2273-2286. https://doi.org/10.1016/j.celrep.2018.07.095.
