Overview
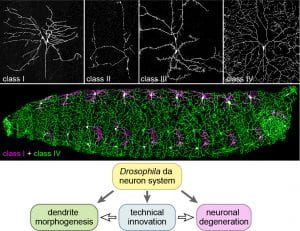
The research goal of the Han lab is to decipher the secrets of the neuronal morphogenesis and neurodegeneration using cutting-edge technologies. Our primary model systems are the beautiful dendritic arborization (da) somatosensory neurons of Drosophila larvae (Figure 1). We welcome motivated graduate students, postdoctoral scholars, and technicians to be part of our team.
For neuronal morphogenesis, we aim to uncover the rules governing the branching patterns of neuronal dendrites at molecular and cellular levels. We would like not only to discover the molecular players that are essential for specific branching patterns of neurons but also to understand how the activity of these players in time and space produces particular branching outcomes.
For neurodegeneration, we are curious about how neurons interact with other cell types during neuronal damage, repair, and degeneration. Our current focus is on phagocytic interactions between neurons and phagocytes resulting from exposure of the “eat-me” signal phosphatidylserine (PS) on neurons. We showed that PS is responsible for inducing engulfment of neurons by resident phagocytes in various neurodegenerative contexts and that PS-induced phagocytosis can act as a driver for neurodegeneration in injury and under certain genetic conditions. We are currently investigating the regulation of PS exposure on neurons and the recognition of PS by phagocytes.
To address the above questions, we are keen on developing novel genetic, molecular, and cellular tools that can enable us to dissect biological processes with greater power and flexibility (Figure 1). One of our focuses is on the development of CRISPR and optogenetic tools in Drosophila; another is on novel microscopy techniques and quantitative image analyses.
I. Dendrite morphogenesis
Dendrite morphogenesis sets the foundation for neural circuit formation. A neuron’s potential for dendrite growth and branching is determined by the intrinsic genetic program that gives rise to the neuron’s unique identity. During early phases of dendrite morphogenesis, a neuron develops a dendritic arbor with a particular size, shape, and branching pattern. The dendritic arbor is then maintained over time to carry out its function in information collection and processing. Dendrite arborization takes place in a complex cellular environment in which neurons interact with many other cells. We are interested to discover the genetic programs and neuron-environment interactions that shape every step of dendrite morphogenesis.
(1) Towards understanding how extracellular environments regulate dendrite morphogenesis, we discovered that heparan sulfate proteoglycans (HSPGs), extracellular glycoproteins carrying heparan sulfate chains, provide permissive growth signals specifically to space-filling neurons (Poe et al, 2017). Using LarvaSPA, a novel long-term timelapse imaging method we invented (Ji and Han, 2020), we showed that HSPGs are not required for extension or branching of high-order dendrites, but rather stabilize dynamic dendritic branches by promoting microtubule stabilization (Movie 1). In addition, we found that Ptp69D, a putative membrane receptor carrying tyrosine phosphatase activity, promotes the growth of high order dendrites of space-filling neurons. Our current goal in this project is to determine the nature of the permissive signals and to dissect how Ptp69D promotes dendrite branching using novel quantitative tools.
Movie 1. Dendrite dynamics in HSPG deficiency
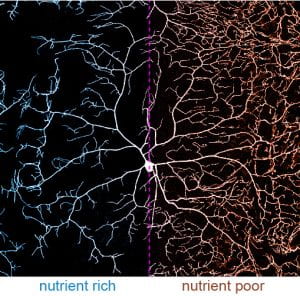
(2) At the systemic level of neuronal growth regulation, we discovered that the environmental nutrient condition profoundly impacts dendrite morphology (Poe et al., 2020). Some neurons exhibit dynamic dendrite growth during the entire larval development. Interestingly, their dendrite growth is “spared” under nutrient deficiency. In other words, these neurons grow faster relative to neighboring non-neural tissues, resulting in dendrite overgrowth (Figure 2). This dendrite overgrowth heightens animal responses to sensory stimuli, possibly providing animals a survival advantage under environmental challenges. Our work reveals the underlying molecular mechanism that involves the master growth-regulating Tor pathway, autophagy, and the stress sensor FoxO. Our future goal in this project is to characterize other effectors that are involved in dendrite sparing.
(3) In addition, we have several other exciting ongoing projects related to dendrite morphogenesis. First, we are using a novel optogenetic approach to investigate how the cytoskeleton is regulated globally in the whole arbor and locally in individual branches to control dendrite dynamics. Second, we are investigating how particular neuronal identities are maintained by specific genetic programs in neurons. Third, we are studying how different classes of neurons interact with one another to allow repulsion or coexistence. Lastly, we are conducting genome-wide mosaic-based screens based on our MAGIC system (Allen et al., 2021) to search for genes important for dendrite patterning.
II. Neurodegeneration
Eat-me signal: In both the central nervous system and the peripheral nervous system, resident phagocytes engulf dead neurons and degenerating parts of neurons, keeping the nervous system free of neuronal debris resulting from neurodegeneration. Phagocytes accomplish this task by recognizing specific “eat-me” signals on the surface of their targets. By developing an in vivo system for visualizing phosphatidylserine (PS) exposed on da neurons in intact live animals, we showed that PS is specifically exposed on degenerating dendrites in multiple contexts, including neuronal injury (Movie 2), dendrite pruning, and genetically induced dendrite degeneration (Sapar et al., 2018; Ji et al., 2022). By ectopically inducing PS exposure through genetic manipulations of PS flippases (which prevent PS exposure) and lipid scramblases (which expose PS), we showed that PS exposure on neurons dominantly cause phagocytes to attack neurons, resulting in engulfment-dependent dendrite and axon degeneration (Sapar et al., 2018).
Movie 2. PS exposure on injured dendrites
Regulation of PS exposure: PS-mediated phagocytosis is not merely a subsequent step after neuronal death and neurite fragmentation. We showed that PS-mediated phagocytosis drives dendrite degeneration induced by injury and by genetic disruptions of NAD+ biosynthesis (Ji et al., 2022). Our results suggest that PS asymmetry on neuronal membranes is maintained by factors regulated by local NAD+ levels in dendrite branches. One of our current goals is to determine the identities of these factors and to understand how they are regulated by NAD+. Besides NAD+, we found that PS asymmetry on neurons are maintained by lipid transporters including the PS flippase ATP8A (Sapar et al., 2018) and an ABCA transporter called Eato (Chen et al., 2025). Interestingly, these lipid transporters not only protect neurons but allow phagocytes to recognize PS exposed on neurons more readily by maintaining PS homeostasis in both neurons and phagocytes.
Detection of PS exposure: Once exposed on the cell surface, PS has to be detected by engulfment receptors expressed by nearby phagocytes. We showed that this detection is mediated by Orion, a recently discovered extracellular protein (Ji et al., 2023). Orion serves as a permissive factor to bridge the interaction between PS and the engulfment receptor. Interestingly, the level of extracellular Orion determines the sensitivity of phagocytes to neuronal PS exposure. Our current focus on this project is to characterize the molecular mechanism of Orion’s interactions with PS and the engulfment receptor.
By developing the first in vivo visualization system for neuronal PS exposure in Drosophila, we surprisingly found that some neuronal types naturally expose PS on specific neuronal compartments without inducing phagocytosis, suggesting that PS exposure may play roles beyond neurodegeneration. A major interest of our lab in the next a few years is to understand how this natural PS exposure is regulated and what are the biological functions of the natural PS exposure. This line of investigation will likely illuminate the little-known world of in vivo neuronal PS exposure.
III. Technical innovation
We have a constant interest in developing more powerful approaches for manipulating gene functions and for quantitative microscopy and analysis. These new methods allow us to tackle our problems in ways not possible previously.
(1) Tissue-specific gene knockout by CRISPR: We developed CRISPR-TRiM, a simple, yet powerful, CRISPR-based system to achieve tissue-specific gene knockout in Drosophila (Poe et al., 2019). The key to the high efficiency of this system was optimizations of both Cas9 and gRNA designs for both germline and somatic tissues. We developed a series of gRNA vectors for diverse purposes, including labeling cells carrying CRISPR-generated mutations (Koreman et al, 2021). This approach can be used orthogonally with any other binary system and has been important in all of our ongoing projects. (See Han lab reagents)
(2) Mosaic analysis with MAGIC: We developed a novel CRISPR-based method called MAGIC (mosaic analysis by gRNA-induced crossing-over) for mosaic analysis (Allen et al., 2021). This technique does not rely on site-specific recombination systems and thus can be used with any Drosophila strain (such as wildly derived strains) without the need for genetic modification. We are currently building MAGIC tools for all Drosophila chromosomal arms, which will be used to conduct genome-wide forward genetic screens and be distributed to the Drosophila community. (See Han lab reagents)
(3) We developed an optogenetic system called OptoTrap to manipulate activity of endogenous proteins in Drosophila by light (Xu et al., 2024). Using this approach, we would like to shut off proteins important for dendrite morphogenesis or neurodegeneration whenever and wherever we want (such as in a specific dendrite branch). This approach will provide us unprecedented spatial and temporal resolutions to dissect the molecular processes governing dendrite branching morphogenesis. Important for this type of quantitative manipulations, we have already developed LarvaSPA, a method of long-term time-lapse imaging for da neurons (Ji and Han, 2020), and methods of quantitative analysis of dendrite dynamics.
